Projects
Epigenetic gene regulation
Our interest in epigenetics originated in our studies on MEDEA (MEA), which is connected to epigenetic processes at two levels: it is regulated by genomic imprinting and encodes a histone methyltransferase, a subunit of Polycomb Repressive Complex 2 (PRC2). Our studies on the regulation of MEA by genomic imprinting and the molecular function of MEA in the seed continue. Albeit a role for MEA in the embryo had previously been dismissed, using an elegant genetic trick, we could unambiguously demonstrate that MEA is required for embryonic pattern formation in plants as it is in animals. Thus, the role of PRC2 in regulating cell proliferation and patterning is conserved between animals and plants, illustrating convergent evolution in these lineages. In recent years, we have expanded our epigenetics research to other areas, and concomitantly developed a series of tools and bioinformatics workflows to analyze epigenetic modifications, e.g., to quantify the variation in DNA-methylation among cell types, individuals, or populations.
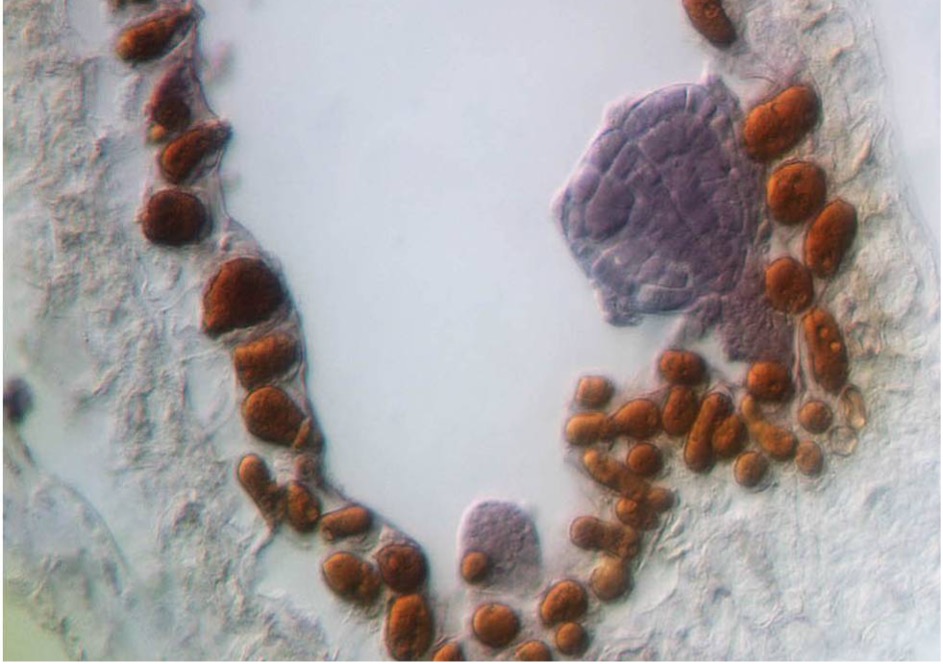 Image by Ueli Grossniklaus
Image by Ueli Grossniklaus
Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB (1998). Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science 280, 446-50. doi: https://doi.org/10.1126/science.280.5362.446.
Wöhrmann HJ, Gagliardini V, Raissig MT, Wehrle W, Arand J, Schmidt A, Tierling S, Page DR, Schöb H, Walter J, Grossniklaus U (2012). Identification of a DNA methylation-independent imprinting control region at the Arabidopsis MEDEA locus. Genes Dev 26, 1837-1850. doi: https://doi.org/10.1101/gad.195123.112.
Pires ND, Bemer M, Müller LM, Baroux C, Spillane C, Grossniklaus U (2016). Quantitative genetics identifies cryptic genetic variation in the paternal regulation of seed development. PLoS Genet 12, e1005806. https://doi.org/10.1371/journal.pgen.1005806.
Simonini S, Bemer M, Bencivenga S, Gagliardini V, Pires ND, Desvoyes B, van der Graaff E, Gutierrez C, Grossniklaus U (2021). The Polycomb group protein MEDEA controls cell proliferation and embryonic patterning in Arabidopsis. Dev Cell 56, 1945-1960. https://doi.org/10.1016/j.devcel.2021.06.004.
Kartal Ö, Schmid MW, Grossniklaus U (2020). Cell type-specific genome scans of DNA methylation diversity indicate an important role for transposonable elements. Genome Biol 21, 172. https://doi.org/10.1186/s13059-020-02068-2.
Cell-specification in reproductive development
We contributed significantly to the establishment of new technologies that allow the genome-wide study of specific cell types that are deeply embedded in surrounding tissues. This includes the use of laser-assisted microdissection to collect individual cell types from tissue sections and to perform transcriptome and methylome studies from minute amounts of DNA or RNA extracted from them,Using this method, we established the first cell type-specific expression maps of reproductive tissues, including the cells of the embryo sac. We also performed RNAseq experiments on specific cell types in different mutants or accessions, e.g., comparing egg cells in apomictic and sexual plants, providing unprecedented insights into the molecular networks controlling these processes. Using such technologies, we identified transcription factors of the RKD family that play a key role in the specification of egg cell fate and found a paternally delivered cyclin that initiates endosperm development.
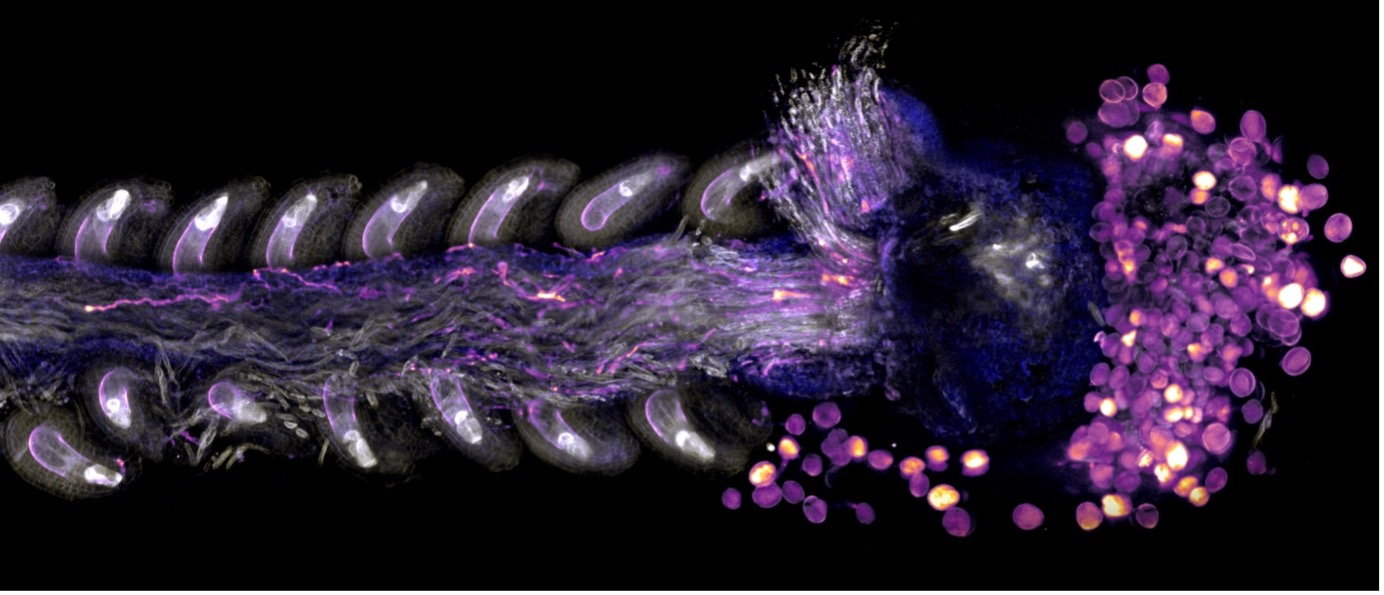 Image by Nicholas Desnoyer
Image by Nicholas Desnoyer
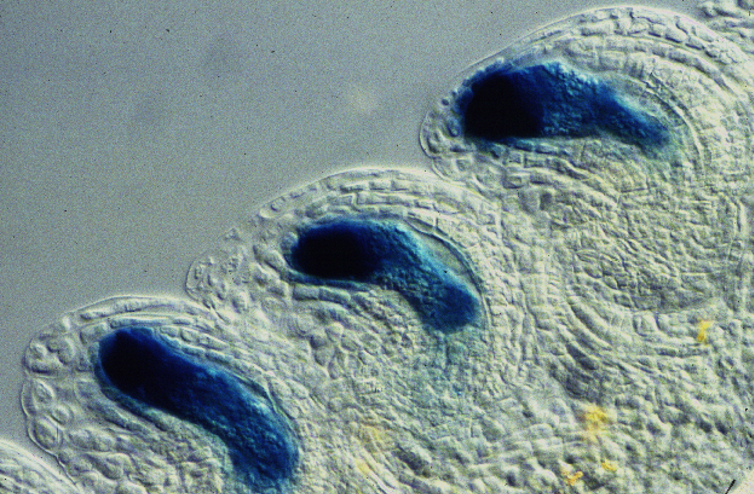 Image by Ueli Grossniklaus
Image by Ueli Grossniklaus
Wuest SE, Vijverberg K, Schmidt , Weiss M, Gheyselinck J, Lohr M, Wellmer F, Rahnenführer J, von Mering C, Grossniklaus U (2010). Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol 20, 506-512. https://doi.org/10.1016/ j.cub.2010.01.051.
Koszegi D, Johnston AJ, Rutten T, Czihal A, Altschmied L, Kumlehn J, Wüst SE, Kirioukhova O, Gheyselinck J, Grossniklaus U, Bäumlein H (2011). Members of the RKD transcription factor family induce an egg cell-like gene expression program. Plant J 67, 280-291. doi: https://doi.org/10.1111/j.1365-313X.2011.04592.x.
Florez-Rueda AM, Paris M, Schmidt A, Widmer A, Grossniklaus U, Städler T (2016). Genomic imprinting in the endosperm is systematically perturned in abortive hybrid tomato seeds. Mol Biol Evol 33, 2935-2946. https://doi.org/10.1093/molbev/msw175.
Simonini S, Bencivenga S, Grossniklaus U (2024). A paternal signal induces endosperm proliferation upon fertilization in Arabidopsis. Science 383, 646-653. doi: https://doi.org/10.1126/science.adj4996.
Cell-cell communication – the FERONIA pathway
Our studies of cell-cell communication during fertilization yielded more general insights into the novel CrRLK1L signal transduction pathway in plants, playing diverse roles in development, physiology, and interactions with the environment. Using a combination of genetic and molecular approaches, we dissected the FERONIA (FER) and ANXUR1/2 signal transduction pathways and identified several novel components, including LRX proteins providing a link to the regulation of cell wall properties. Moreover, we could recently show that FER to be the first known species-specific recognition key during fertilization, unravelling an important mechanism for gametophyte recognition and speciation. We have extended our studies on the FER pathway from Arabidopsis to Marchantia, highlighting the importance of this pathway in controlling cell expansion.
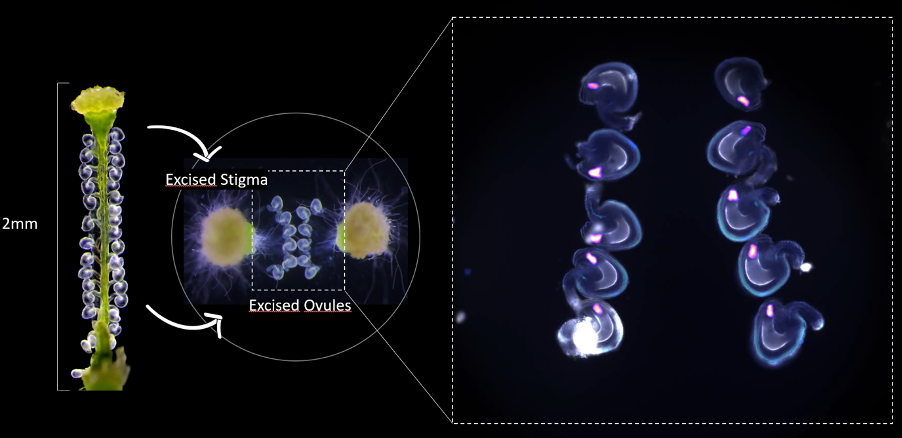
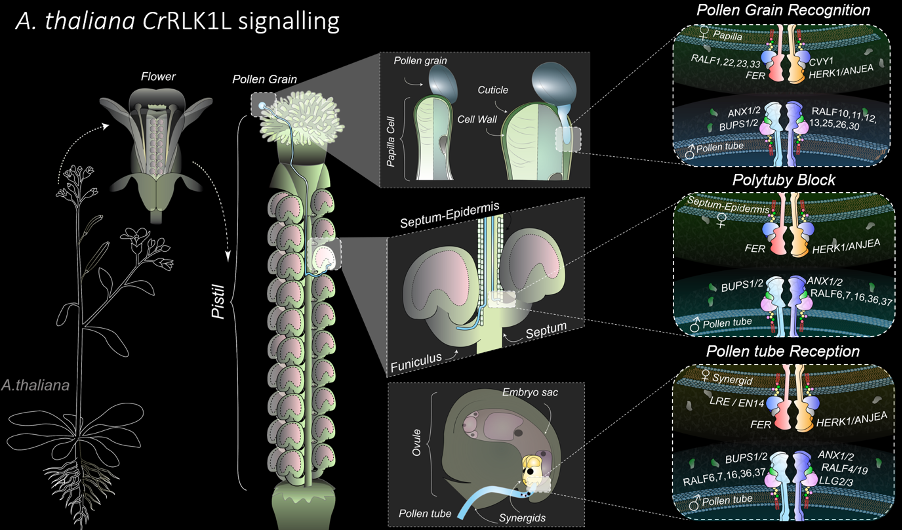
- Image and Illustration by Nicholas Desnoyer
Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U (2007). The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317, 656-660. doi: https://doi.org/10.1126/science.1143562.
Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U (2010). Conserved molecular components for pollen tube reception and fungal invasion. Science 330, 968-971. doi: https://doi.org/10.1126/science.1195211.
Mecchia M, Santos-Fernandez G, Duss NN, Somoza SC, Boisson-Dernier A, Gagliardini V, Martínez-Bernardini A, Ndinyanka Fabrice T, Ringli C, Muschietti JP, Grossniklaus U (2017). RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 358, 1600-1603. https://doi.org/10.1126/science.aao5467.
Moussu S, Broyart C, Santos-Fernandez G, Augustin S, Wehrle S, Grossniklaus U, Santiago J (2020). Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth. Proc Natl Acad Sci USA 117, 7494-7503. https://doi.org/10.1073/pnas.2000100117.
Analysis of Plant Genomes
The analysis of genomes, both of model and non-model species, has become increasingly important, both for functional and evolutionary studies. We exploit natural variation to gain insights into the developmental processes we are interested in as well as to develop genomic resources for functional studies. We have been involved in several genome projects to which we contributed through the characterization of gene families or chromosome-scale assemblies based on chromosome conformation information we obtained by Hi-C. Importantly, we have developed bioinformatic tools to assemble haplotype-aware genomes of highly heterozygous species, which are extremely difficult to assemble.
 Image by Alejandro Giraldo Fonseca
Image by Alejandro Giraldo Fonseca
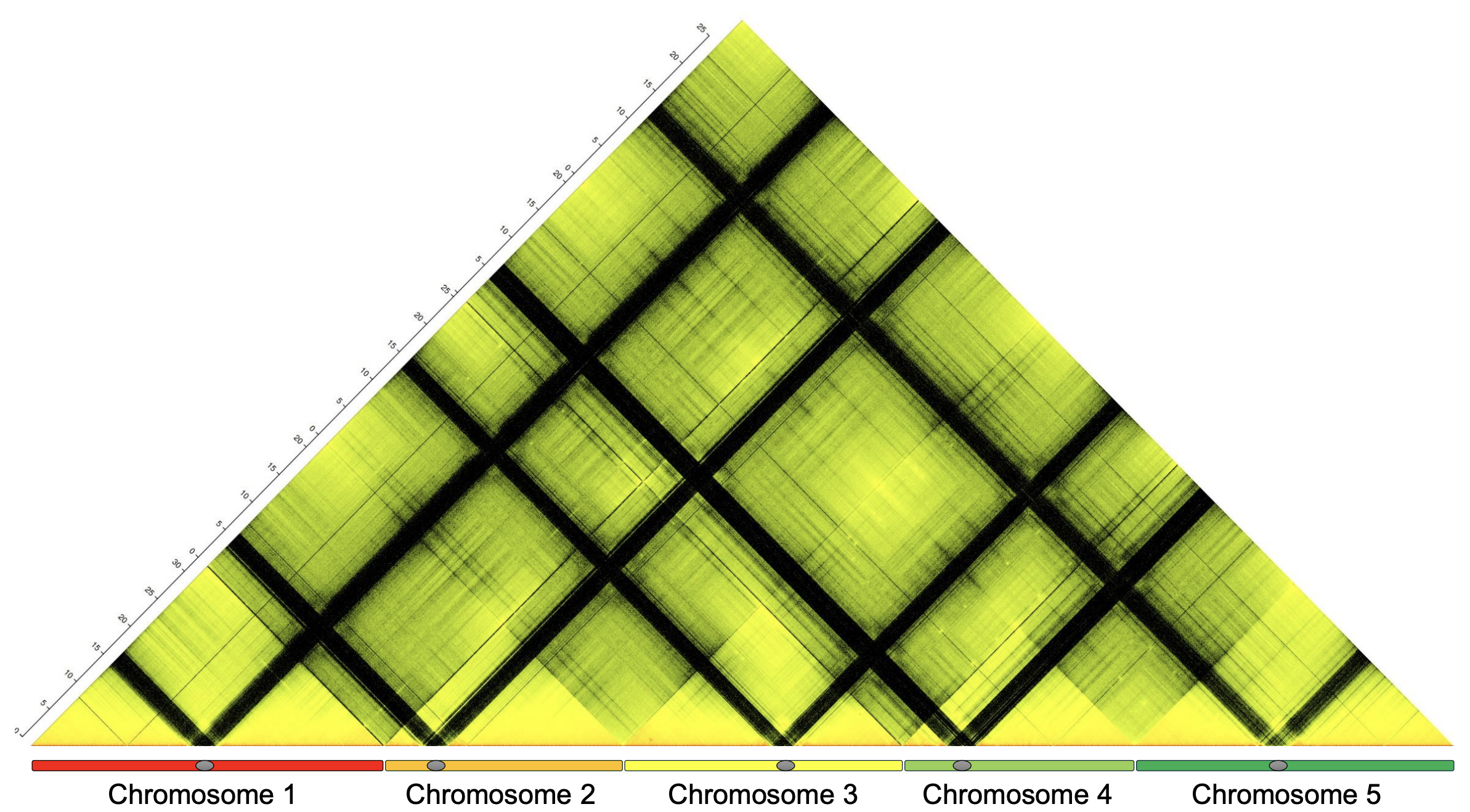 Image by Victor Mac
Image by Victor Mac
Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, et al. (2017). Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287-304. https://doi.org/10.1016/ j.cell.2017.09.030.
Kliver S, Rayko M, Komissarov A, Bakin E, Zhernakova D, Prasad K, Rushworth C, Baskar R, Smetanin D, Schmutz J, Rokhsar DS, Mitchell-Olds T, Grossniklaus U, Brukhin V (2018). Assembly of the Boechera retrofracta genome and evolutionary analysis of apomixis-associated genes. Genes 9, 185. https://doi.org/10.3390/genes9040185.
Zhang L, Cai X, Wu J, Liu M, Grob S, Cheng F, Liang J, Cai C, Liu Z, Liu B, Wang F, Li S, Liu F, Li X, Cheng L, Yang W, Li MH, Grossniklaus U, Zheng H, Wang X (2018). Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic Res 5, 50. https://doi.org/10.1038/s41438-018-0071-9.
Kuon JE, Qi W, Schläpfer P, Hirsch-Hoffmann M, von Bieberstein PR, Patrignani A, Poveda L, Grob S, Keller M, Shimizu-Inatsugi R, Grossniklaus U, Vanderschuren H, Gruissem W (2019). Haplotype-resolved genomes of geminivirus-resistant and geminivirus-sisceptible African cassava cultivars. BMC Biol 17, 75. https://doi.org/10.1186/s12915-019-0697-6.
Engineering of Apomixis
One of our long-standing interests is apomixis, the production of clonal seed, which bears great potential for agriculture. In Arabidopsis and maize, we used genetic and molecular approaches to identify mutations that either prevent meiosis or induce parthenogenesis. When such mutations are combined, it is expected to lead to the production of clonal progeny. Using these genetic tools, we were successful to produce the first clonal progeny in a crop, i.e., maize. We have also performed studies in natural apomicts to better understand the ecological role of clonal reproduction and to demonstrate the heritability of phenotypes across apomictic generations. We currently participate in a large, international consortium with the goal to engineer apomixis in crop species that are relevant to Sub-Sharan Africa to benefit of smallholder farmers.
 Images by Nina Chumak
Images by Nina Chumak
Spillane C, Curtis MD, Grossniklaus U (2004). Apomixis technology development-virgin births in farmers' fields? Nat Biotechnol 22, 687-691. doi: https://doi.org/10.1038/nbt976.
Sailer C, Schmid B, Grossniklaus U (2016). Apomixis Allows the Transgenerational Fixation of Phenotypes in Hybrid Plants. Curr Biol 26, 331-337. doi: https://doi.org/10.1016/j.cub.2015.12.045.
Fox TW, Albertsen MC, Williams ME, Lawit SJ, Chamberlin MA, Grossniklaus U, Brunner GA, Chumak N, Bernardes de Asis J, Pasquer F (2016). Methods and compositions for the production of unreduced, non-recombined gametes and clonal offspring. US Patent Application US2016/0031271W. https://patents.justia.com/patent/20180142251.
Brukhin V, Osadtchiy JV, Florez-Rueda AM, Smetanin D, Bakin E, Nobre MS, Grossniklaus U (2019). The Boechera genus as a resource for apomixis research. Frontiers Plant Sci 10, 392. https://doi.org/ 10.3389/fpls.2019.00392.
The Mechanics of Growth
In recent years, we developed a strong interest to better understand how mechanical forces control cellular growth and morphogenesis. Cells embedded in tissues have to coordinate their growth with their neighbors, and physical forces play an important role in cell and organ morphogenesis. Yet, tools to measure such forces in living cells are scarce, so together with engineers, material scientists, and physicists, we set out to develop experimental approaches to measure the minute forces relevant to living cells and tissues.
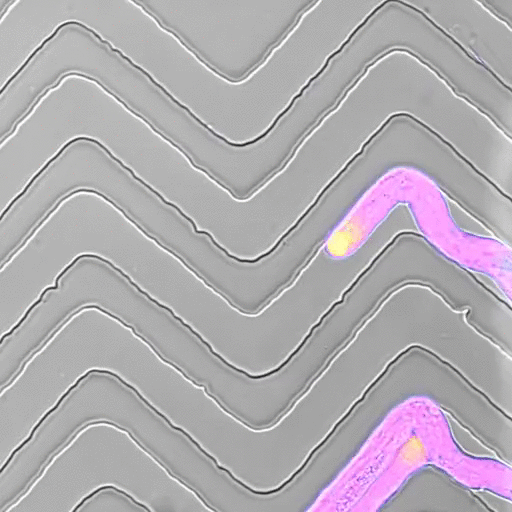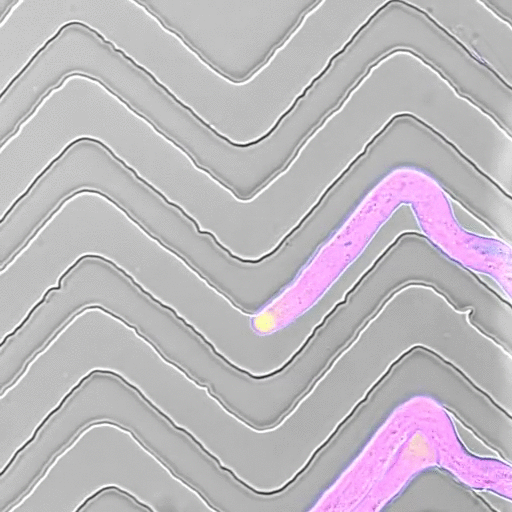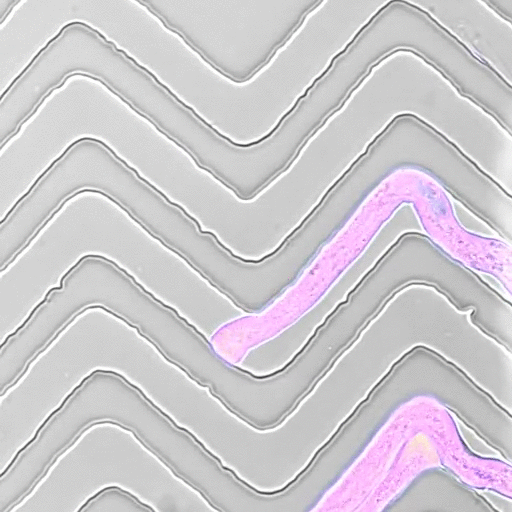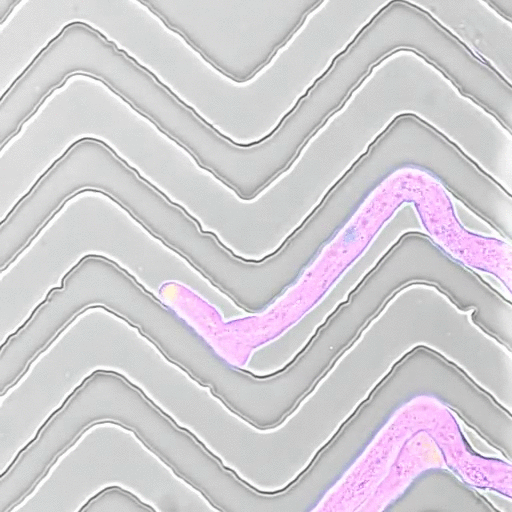
- Movie by Marta Belloli
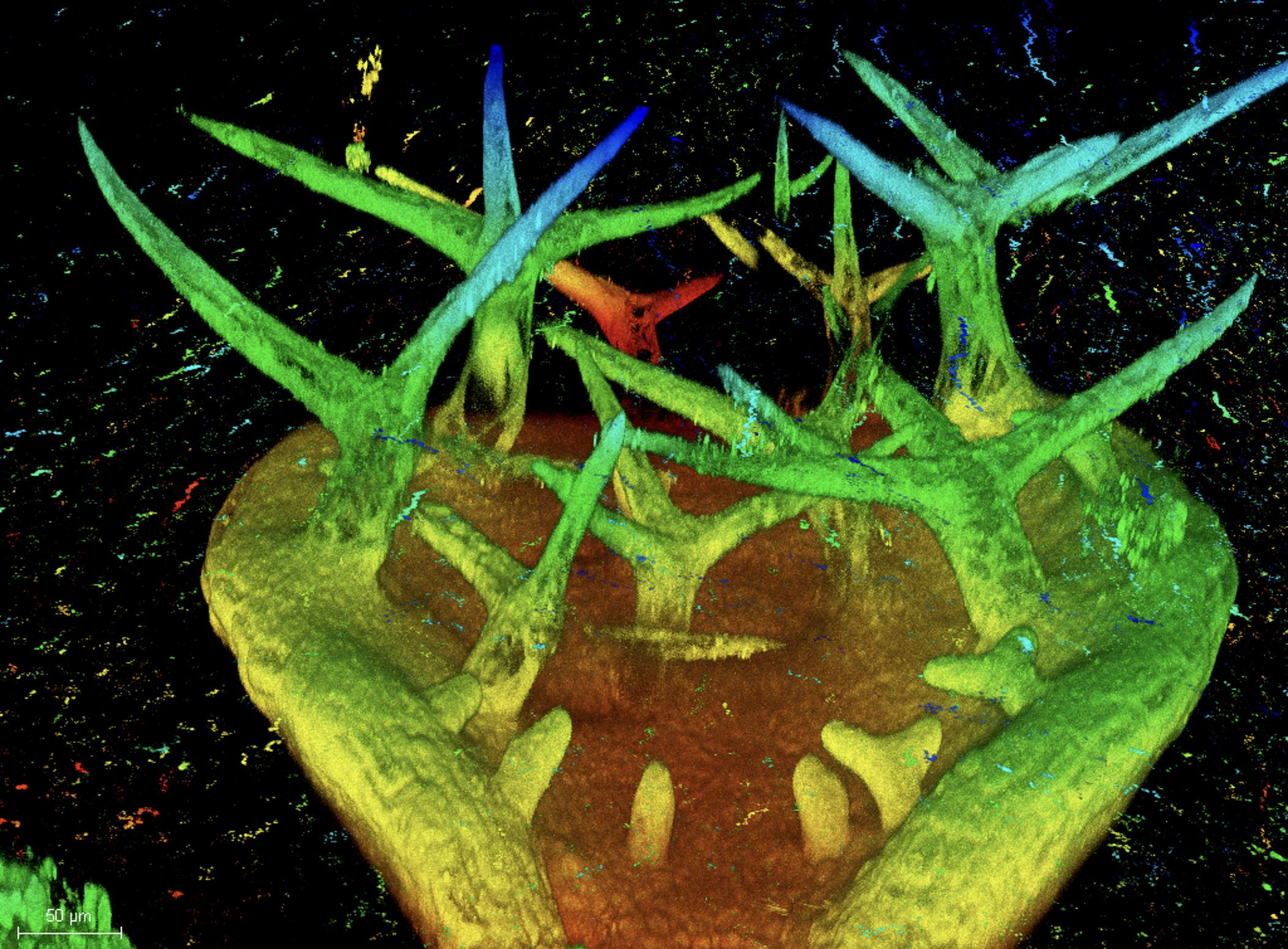
- Image by Dorothee Stöckle
Vogler H, Draeger C, Weber A, Felekis D, Eichenberger C, Routier-Kierzkowska AL, Boisson-Dernier A, Ringli C, Nelson BJ, Smith RS, Grossniklaus U (2013). The pollen tube: a soft shell with a hard core. Plant J 73, 617-627. doi: https://doi.org/10.1111/tpj.12061.
Burri JT, Vogler H, Läubli NF, Hu C, Grossniklaus U, Nelson BJ (2018). Feeling the force: how pollen tubes deal with obstacles. New Phytol 220, 187-195. https://doi.org/10.1111/nph.15260.
Burri JT, Saikia E, Läubli NF, Vogler H, Wittel FK, Rüggeberg M, Herrmann HJ, Burgert I, Nelson BJ, Grossniklaus U (2020). A single touch can provide sufficient mechanical stimulation to trigger Venus flytrap closure. PLoS Biology 18, e3000740. https://doi.org/10.1371/journal.pbio.3000740.
Läubli NF, Burri JT, Marquard J, Vogler H, Mosca G, Vertti-Quintero N, Shamsudhin N, deMello A, Grossniklaus U, Ahmed D, Nelson BJ (2021). 3D mechanical characterization of single cells and small organisms using acoustic manipulation and force microscopy. Nat Commun 12, 2583. doi: https://doi.org/10.1038/s41467-021-22718-8.
Subtropical crops – Marama & Papaya
Subtropical crops play an important role in local agronomical systems but only few of them are traded internationally. Consequently, they receive little attention in research, breeding, or improvement of agricultural practices to produce them. Furthermore, a series of subtropical plants would be suited as a crop but are not cultivated. One such ‘orphan crop’ is the highly nutritious African legume Tylosema fassoglense, known by the common name Marama bean or Tamani berry. This species has a very high drought tolerance, making it potentially attractive for cultivation in times of climate change. Together with South-African researchers, we are aiming at establishing an annotated reference genome of Marama bean as a basis for future research. In another line of work relevant to climate change, we are studying environmentally induced sex reversal in Papaya carica, which reduces the harvests of smallholder famers and producers.
 Image by Tiago Meier
Image by Tiago Meier
 Images by Jonata Ribeiro
Images by Jonata Ribeiro
Jackson JC, Duodu KG, Holse M, Lima de Faria MD, Jordaan D, Chingwaru W, Hansen A, Cencic A, Kandawa-Schultz M, Mpotokwane SM, Chimwamurombe P, de Kock HL, Minnaar A (2010). The morama bean (Tylosema esculentum): a potential crop for southern Africa. Adv Food Nutr Res 61, 187-246. https://doi.org/10.1016/B978-0-12-374468-5.00005-2.
Tadele Z (2019). Orphan crops: their importance and the urgency of improvement. Planta 250, 677-694. https://doi.org/10.1007/s00425-019-03210-6.
Epigenetic variation in ecology and evolution
In plants, epigenetic changes can be heritable over generations and occur more frequently than genetic ones. Thus, epigenetic variation may allow rapid responses and has the potential to play a key role in the adaptation to environmental change. Epigenetic variation, which leads to (meta)stable changes in gene expression without changes in the DNA sequence, arises about 10’000 times more often than genetic variation and has the potential to occur in two states depending on the environment, which could help in the adaptation of crops to a changing climate. So far, the ecological and evolutionary significance of epigenetic variation is largely unknown. Over the last years, we have collected experimental data demonstrating a role for epigenetic variation in the adaptation to changes in the biotic and abiotic environment using Arabidopsis as a model. We are currently studying the stability and heritability of epigenetic variation in lab and field.
 Image by Alex Plüss
Image by Alex Plüss
Fakheran S, Paul-Victor C, Heichinger C, Schmid B, Grossniklaus U, Turnbull LA (2010). Adaptation and extinction in experimentally fragmented landscapes. Proc Natl Acad Sci USA 107, 19120-19125. https://doi.org/10.1073/pnas.1010846107.
Paszkowski J, Grossniklaus U (2011). Selected aspects of transgenerational epigenetic inheritance and resetting in plants. Curr Opin Plant Biol 14, 195-203. doi: https://doi.org/10.1016/j.pbi.2011.01.002.
Hirsch S, Baumberger R, Grossniklaus U (2012). Epigenetic variation, inheritance, and selection in plant populations. Cold Spring Harb Symp Quant Biol 77, 97-104. doi: https://doi.org/10.1101/sqb.2013.77.014605.
Schmid MW, Heichinger C, Coman Schmid D, Guthörl D, Gagliardini V, Bruggmann R, Aluri S, Aquino C, Schmid B, Turnbull LA, Grossniklaus U (2018). Contribution of epigenetic variation to adaptation in Arabidopsis. Nat Commun 9, 4446. https://doi.org/10.1038/s41467-018-06932-5.
Evo-Devo: Marchantia polymorpha
A central question in biology is how the tremendous variation in the morphology of organisms arose in evolution. While it was previously thought that different genes control morphological differences between organisms, it is now widely accepted that similar genes control similar traits across species. The evolution of land plants from an algal ancestor some 470 million years ago is characterized by the innovation of various traits, e.g., the evolution of a water conducting tissue that was crucial for the success on land. We study the liverwort Marchantia polymorpha as early diverging land plant lineage that has similar types of gene families as higher plants, suggesting that the differences in development evolved by co-opting and modifying existing developmental programs and genetic networks, rather than through the evolution of novel genes.
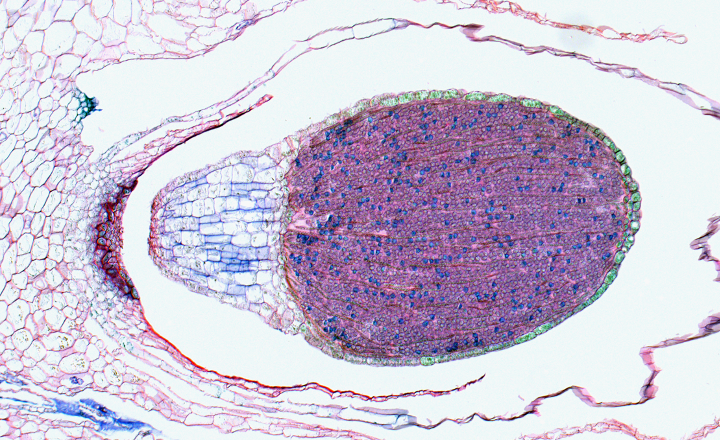 Image by Tom Dierschke
Image by Tom Dierschke
 Movie by Nicholas Desnoyer
Movie by Nicholas Desnoyer
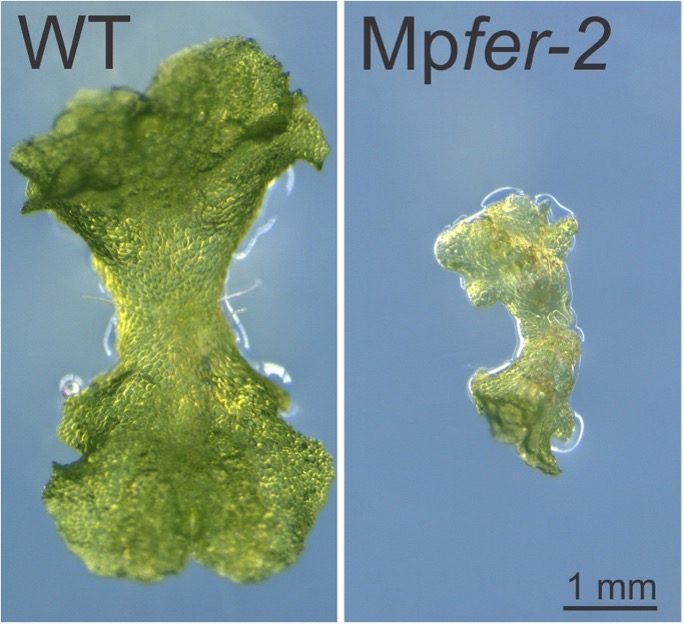 Image by Martin Mecchia
Image by Martin Mecchia
Rövekamp M, Bowman JL, Grossniklaus U (2016). Marchantia MpRKD regulates the gametophyte-sporophyte transition by keeping egg cells quiescent in the absence of fertilization. Curr Biol 26, 1782-1789. doi: https://doi.org/10.1016/j.cub.2016.05.028.
Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, et al. (2017). Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287-304. https://doi.org/10.1016/ j.cell.2017.09.030.
Schmid MW, Giraldo-Fonseca A, Rövekamp M, Smetanin D, Bowman JL, Grossniklaus U (2018). Extensive epigenetic reprogramming during the life cycle of Marchantia polymorpha. Genome Biol 19, 9. https://doi.org/10.1186/s13059-017-1383-z.
Mecchia MA, Rövekamp R, Giraldo-Fonseca A, Meier D, Gadient P, Voglere H, Limacher D, Bowman JL, Grossniklaus U (2022). The single Marchantia polymorpha FERONIA homolog reveals an ancestral role in regulating cellular expansion and integrity. Development 149, dev200580. https://doi.org/ 10.1242/dev.200580.
Community ressources
UGOmics
UGomics Server is an interactive Shiny-based platform developed at the Grossniklaus Lab (by Alejandro Giraldo-Fonseca) to streamline access to bioinformatics tools and resources. The platform offers organism-specific portals for Arabidopsis thaliana, Marchantia polymorpha, and many more, featuring in-house RNA-seq expression servers, external genomic databases, and specialized analysis tools. It also highlights tool contributions from lab members, facilitating tool sharing and accessibility. UGomics simplifies genomic research by integrating key resources into a user-friendly interface, thought and made for efficient bioinformatics workflows.
The information and tools are classified by:
- Organism
- Type of data analysis
- People who made the tool and/or have experience with a specific data analysis
You can visit the repository here UGomics
FRET-IBRA
Downloads:
The source files, examples and tutorial are hosted on github:
- Source
- Tutorial & examples
- Operating Systems: Linux/Mac OS (64-bit)
Archive:
Rcount
Downloads:
The user guide, source and binary files are now hosted in github:
- User guide
- Linux (64-bit)
- Windows (64-bit)
- Mac (64-bit)
- Source
Data:
HiCdat
Downloads:
The user guide, source and binary files are now hosted in github:
- User guide
- Linux (64-bit)
- Mac (64-bit)
- Windows (64-bit)
- Source
Data:
Arabidopsis thaliana (pre-process tutorial)
Citation:
Grob S, Schmid MW, Grossniklaus U. (2014) HiC Analysis in Arabidopsis Identifies the KNOT, a Structure with Similarities to the flamenco Locus of Drosophila. Molecular Cell, 55(5): 678-693 [PubMed]

