Projects
Functional genomics to understand beta-rhizobial symbiosis and host specificity
Funding: SNF project 310030_215282 (2023-2027)
People involved: Dr. Paula Bellés Sancho, Dr. Yilei Liu, Martinus Hjorungnes, Romy Leemann
Collaborators: Dr. Christian Ahrens (Agroscope-Molecular Ecology and Swiss Institute of Bioinformatics), Prof. Nicola Zamboni (IMSB, ETHZ), Dr. Euan K James (The James Hutton Institute, Dundee, UK).
The successful interaction between specific rhizobia and host plants lead to the formation of specialized symbiotic organs, called root nodules, in which the bacteria fix atmospheric nitrogen (N2) and make it available to the plant. In exchange, the plant provides carbon and energy sources generated by photosynthesis. A complex series of events, coordinated by both plant and bacterial signals, underlies the development of this symbiotic interaction1. Until recently, rhizobia were thought to belong exclusively to the alpha-subclass of proteobacteria (alpha-rhizobia, isolated first in 1888). This changed in 2001, when two genera belonging to the beta-proteobacteria (Burkholderia and Cupriavidus) were discovered (beta-rhizobia)2. For the legume nodulating strains belonging to the environmental clade of the genus Burkholderia, recently a new genus, Paraburkholderia, has been proposed. The strain Paraburkholderia phymatum STM815T stands out for its ability to establish a nitrogen-fixing symbiosis with more than 50 different legumes (including mimosoid and papilionoid) as well as to reduce nitrogen in free-living conditions (Figure 1a,b)3–8. Moreover, the availability of the complete genome sequence makes it an ideal model to characterize the whole gene/protein expression profile of P. phymatum STM815T grown in different environments including the root nodule5.
In order to understand the molecular mechanism underlying beta-rhizobial symbiosis and identify differences and commonalities between the strategies used by alpha- and beta-rhizobia, we use functional genomic approaches such as transposon sequencing (Tn-Seq), RNA-sequencing, shotgun proteomics and metabolomics (Figure 1c). Our group was the first to determine the whole transcriptome profile of P. phymatum STM815T under nitrogen limitation (a condition that partially mimics the condition encountered by free-living rhizobia in nitrogen-starved soils) and in symbiosis with the economically important papilionoid legume Phaseolus vulgaris (common bean)6. Recently, the combination of metabolomics and dual RNA-sequencing on nodules allowed us to identify novel regulatory pathways in P. phymatum STM815T such as the repression of the production of indole-3-acetic acid (IAA) by the master activator of nitrogen-fixation NifA9.
The goal of this project is to identify and characterize beta-rhizobial genes important for i) bacterial competition in the soil, ii) symbiotic partner recognition, iii) symbiosis establishment, iv) N2-fixation and v) nodule senescence.
Figure 1. a) Common bean root-nodules induced and colonized by P. phymatum STM815T. b) Section of common bean nodules colonized by P. phymatum, plant cell walls stained with calcofluor white (green) and bacterial cells with Syto9 DNA (magenta) (picture from Bellés-Sancho et al., 2022). c) Functional genomic approaches used in our laboratory to study rhizobial symbiosis.
References:
- Bellés-Sancho, P., Beukes, C., James, E. K. & Pessi, G. Nitrogen-fixing symbiotic Paraburkholderia species: current knowledge and future perspectives. Nitrogen 4, 135–158 (2023).
- Moulin, L., Munive, A., Dreyfus, B. & Boivin-Masson, C. Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature 411, 948–950 (2001).
- Elliott, G. N. et al. Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytologist 173, 168–180 (2007).
- Talbi, C. et al. Burkholderia phymatum strains capable of nodulating Phaseolus vulgaris are present in moroccan soils. Appl Environ Microbiol 76, 4587–4591 (2010).
- Moulin, L. et al. Complete Genome sequence of Burkholderia phymatum STM815T, a broad host range and efficient nitrogen-fixing symbiont of Mimosa species. Stand Genomic Sci 9, 763–774 (2014).
- Lardi, M., Liu, Y., Purtschert, G., de Campos, S. B. & Pessi, G. Transcriptome analysis of Paraburkholderia phymatum under nitrogen starvation and during symbiosis with Phaseolus vulgaris. Genes (Basel) 8, (2017).
- Lardi, M., de Campos, S. B., Purtschert, G., Eberl, L. & Pessi, G. Competition experiments for legume infection identify Burkholderia phymatum as a highly competitive β-rhizobium. Front Microbiol 8, (2017).
- Bellés-Sancho, P. et al. Paraburkholderia phymatum homocitrate synthase NifV plays a key role for nitrogenase activity during symbiosis with papilionoids and in free-living growth conditions. Cells 10, 952 (2021).
- Bellés-Sancho, P. et al. A novel function of the key nitrogen-fixation activator NifA in beta-rhizobia: Repression of bacterial auxin synthesis during symbiosis. Front Plant Sci 13, (2022).
Competitiveness traits of Paraburkholderia strains in the soil
Funding: SNF project 310030_215282 (2023-2027)
People involved: Dr. Paula Bellés Sancho, Kim Bolli, Martinus Hjorungnes, Giada Ornaghi, Dr. Yilei Liu
Rhizobia must endure the harsh conditions encountered in the soil to be able to colonize plant roots. Drought, pH, salinity, temperature, and the presence of toxic substances are some examples of environmental stresses that rhizobia must withstand, as are biotic factors such as the presence of other bacteria species competing for common resources. Therefore, the competitiveness of a rhizobia strain is directly influencing a successful colonization of the plant1.
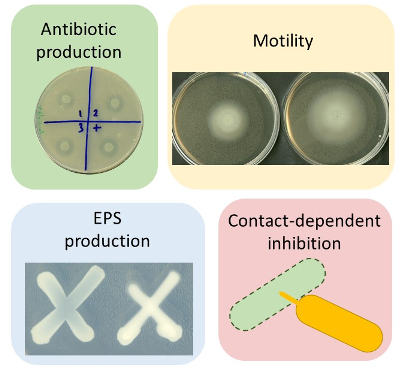
P. phymatum STM815T stands out among the beta-rhizobia group for being the most competitive strain in vitro and for nodulating papilionoid legumes2–4. Our primary goal is to elucidate the strategies and identify traits that P. phymatum STM815T employs to outcompete other strains. In our group we showed that P. phymatum has two types 6 secretion systems (T6SS), one of which contributes to outcompete other beta-rhizobia strains in a contact-dependent manner5,6. Moreover, P. phymatum STM815T produced more exopolysaccharides (EPS) and was more motile compared to other strains, two traits crucial for competitiveness (Figure 1)4. We also propose that the overproduction of the hormone auxin by this strain may enhance the occurrence of infection events and, as result, improve the frequency of root colonization4. Additionally, we recently show that another beta-rhizobial strain, Paraburkholderia sabiae LMG24235, shows T6SS-dependent antagonistic activity towards important plant pathogens such as Pseudomonas syringae and Pectobacterium carotovorum. Moreover, co-inoculation of potato tubers with P. sabiae LMG24235 resulted in a drastic reduction of soft rot caused by P. carotovorum and P. sabiae’s protective effect was partly dependent on T6SS-17.
Figure 1. Phenotypic traits of P. phymatum and P. sabiae that provide a competitive advantage against other bacteria and for root nodulation.
The aim of this project consists in investigating the molecular mechanisms underlying the successful competitiveness of P. phymatum and P. sabiae.
References:
- Bellés-Sancho, P., Beukes, C., James, E. K. & Pessi, G. Nitrogen-fixing symbiotic Paraburkholderia species: current knowledge and future perspectives. Nitrogen 4, 135–158 (2023).
- Elliott, G. N. et al. Burkholderia spp. are the most competitive symbionts of Mimosa, particularly under N-limited conditions. Environ Microbiol 11, 762–778 (2009).
- Moulin, L. et al. Complete Genome sequence of Burkholderia phymatum STM815T, a broad host range and efficient nitrogen-fixing symbiont of Mimosa species. Stand Genomic Sci 9, 763–774 (2014).
- Lardi, M., de Campos, S. B., Purtschert, G., Eberl, L. & Pessi, G. Competition experiments for legume infection identify Burkholderia phymatum as a highly competitive β-rhizobium. Front Microbiol 8, (2017).
- de Campos, S. B., Lardi, M., Gandolfi, A., Eberl, L. & Pessi, G. Mutations in two Paraburkholderia phymatum type VI secretion systems cause reduced fitness in interbacterial competition. Front Microbiol 8, 2473 (2017).
- Hug, S. et al. Differential expression of Paraburkholderia phymatum type VI secretion systems (T6SS) suggests a role of T6SS-b in early symbiotic interaction. Front Plant Sci 12, (2021).
- Hug, S. et al. Paraburkholderia sabiae uses one type VI secretion system (T6SS-1) as a powerful weapon against notorious plant pathogens. Microbiol Spectr Jul, e01622-23 (2023)
Beta-rhizobia’s potential as competitive natural fertilizer under the effects of climate change
Funding: Uniscientia, Project-Nr. 211-2023/Biofertilizers for climate change (2023-2025)
People involved: Dr. Paula Bellés Sancho
With the human population likely to approach 9 billion in the next 30 years, agricultural research is focused on improving food production. However, crop productivity is being hampered by the deterioration of arable land, lack of water, and the ongoing effects of climate change1. Plants, as sessile organisms, cannot escape the effects of climate change on their environment, but they can benefit from soil bacteria and their microbiomes to facilitate their adaptation. Plant growth-promoting rhizobacteria (PGPR) are crucial in this adaptation as they can improve nutrient acquisition and plant development through different mechanisms, including the production of the hormone auxin and the conversion of atmospheric nitrogen (N2) to ammonia2. Nonetheless, the viability of PGPR can be compromised by the consequences of climate change (such as high temperatures or desiccation), prompting the search for PGPR that can withstand abiotic stresses.
Our lab and other international groups have discovered unique characteristics of the beta-rhizobial strain P. phymatum STM815T that make it an excellent model system for studying the influence of climate-induced stresses (Figure 1). In fact, P. phymatum STM815T can nodulate over 50 different types of legumes3,4, outcompetes other beta- and alpha-rhizobia in root-nodulation 4–6, thrives in high temperatures, acidic and nutrient-poor environments 3,4,7–11, and possesses mechanisms used by PGPR to promote plant growth such as auxin production12,13, N2-fixation in symbiosis with legumes and in free-living growth conditions10,14,15, and the ability to synthesize 1-aminocyclopropane-1-carboxylate (ACC) deaminase, an enzyme that lowers plant ethylene levels16.
 Figure 1. Important PGPR traits present in P. phymatum STM815T.
Figure 1. Important PGPR traits present in P. phymatum STM815T.
We will use a highly-saturated library of transposon mutants (Tn-seq) and gene expression studies (RNA-seq) to identify and characterize genes involved in P. phymatum STM815T response to climate change stresses (high temperatures and soil acidity) as well as coping with common bean root exudates produced under the same stress conditions.References:
- Iriti, M. & Vitalini, S. Plant metabolomics in the global scenario of food security: A systems-biology approach for sustainable crop production. Int J Mol Sci 19, (2018).
- Arora, N. K., Verma, M. & Mishra, J. Rhizobial Bioformulations: Past, Present and Future. in Rhizotrophs: Plant Growth Promotion to Bioremediation (ed. Mehnaz, S.) 69–99 (Springer Singapore, 2017). doi:10.1007/978-981-10-4862-3_4.
- Moulin, L. et al. Complete Genome sequence of Burkholderia phymatum STM815T, a broad host range and efficient nitrogen-fixing symbiont of Mimosa species. Stand Genomic Sci 9, 763–774 (2014).
- Lardi, M., de Campos, S. B., Purtschert, G., Eberl, L. & Pessi, G. Competition experiments for legume infection identify Burkholderia phymatum as a highly competitive β-rhizobium. Front Microbiol 8, (2017).
- Elliott, G. N. et al. Burkholderia spp. are the most competitive symbionts of Mimosa, particularly under N-limited conditions. Environ Microbiol 11, 762–778 (2009).
- de Campos, S. B., Lardi, M., Gandolfi, A., Eberl, L. & Pessi, G. Mutations in two Paraburkholderia phymatum type VI secretion systems cause reduced fitness in interbacterial competition. Front Microbiol 8, 2473 (2017).
- dos Reis Junior, F. B. et al. Nodulation and nitrogen fixation by Mimosa spp. in the Cerrado and Caatinga biomes of Brazil. New Phytologist 186, 934–946 (2010).
- Talbi, C. et al. Burkholderia phymatum improves salt tolerance of symbiotic nitrogen fixation in Phaseolus vulgaris. Plant Soil 367, 673–685 (2013).
- Melkonian, R. et al. The geographical patterns of symbiont diversity in the invasive legume Mimosa pudica can be explained by the competitiveness of its symbionts and by the host genotype. Environ Microbiol 16, 2099–2111 (2014).
- Lardi, M., Liu, Y., Purtschert, G., de Campos, S. B. & Pessi, G. Transcriptome analysis of Paraburkholderia phymatum under nitrogen starvation and during symbiosis with Phaseolus vulgaris. Genes (Basel) 8, (2017).
- Hug, S. et al. Differential expression of Paraburkholderia phymatum type VI secretion systems (T6SS) suggests a role of T6SS-b in early symbiotic interaction. Front Plant Sci 12, (2021).
- Bellés-Sancho, P. et al. Metabolomics and dual RNAsequencing on root nodules revealed new cellular functions controlled by Paraburkholderia phymatum NifA. Metabolites 11, 455 (2021).
- Bellés-Sancho, P. et al. A novel function of the key nitrogen-fixation activator NifA in beta-rhizobia: Repression of bacterial auxin synthesis during symbiosis. Front Plant Sci 13, (2022).
- Elliott, G. N. et al. Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytologist 173, 168–180 (2007).
- Bellés-Sancho, P. et al. Paraburkholderia phymatum homocitrate synthase NifV plays a key role for nitrogenase activity during symbiosis with papilionoids and in free-living growth conditions. Cells 10, 952 (2021).
- Klonowska, A., Melkonian, R., Miché, L., Tisseyre, P. & Moulin, L. Transcriptomic profiling of Burkholderia phymatum STM815, Cupriavidus taiwanensis LMG19424 and Rhizobium mesoamericanum STM3625 in response to Mimosa pudica root exudates illuminates the molecular basis of their nodulation competitiveness and symbiotic evolutionary history. BMC Genomics 19, (2018)
Using legumes symbionts for space farming
Funding: Josef Huwyler Ruth Bernet-Engeli Foundation and ESA (I-2022-03207) co-funded Research Idea (2022-2024)
People involved: Daphné Golaz
Collaborators: Prof. Dr. Marcel Egli (IMT, HSLU), Dr. Christian Ahrens (Agroscope-Molecular Ecology and Swiss Institute of Bioinformatics), Prof. Nicola Zamboni (IMSB, ETHZ).
The exploitation of the mutualistic relationship between rhizobia and legumes is a solution to ensure a constant supply in organic nitrogen and an acceptable yield in plants grown in prospective extraterrestrial colonies. To understand the effect of exposure to space-like conditions such as reduced gravity in free-living rhizobia, we are combining classical phenotypic analyses with functional genomics approaches in the beta-rhizobia P. phymatum STM815T grown in an artificial microgravity environment. The integration of data gathered through multiples next-generation sequencing methods, namely Tn-Seq, RNA-Seq, shotgun proteomic and metabolomic analyses, allows us to identify and prioritize P. phymatum STM815T genes and metabolomic pathways that are key for rhizobia thriving in simulated microgravity. Mutants for the genes of interest are constructed and phenotypically characterized in vitro - for growth, cell morphology, resistance to abiotic stresses and secondary metabolites production1 - and in planta with the model legume cowpea for assessment of their symbiotic abilities.
References:
- Golaz, D., Papenfuhs, C.K., Bellés-Sancho, P., Eberl, L., Egli, M., and Pessi, G. RNA-seq analysis in simulated microgravity unveils down-regulation of the beta-rhizobial siderophore phymabactin. npj Microgravity 10, 44 (2024). https://doi.org/10.1038/s41526-024-00391-7